Instructions for
running an R script on NeSI Mahuika
1. Log into NeSI Mahuika via Mobaxterm (as per steps 16 and 17 found here).
2. At the Mobaxterm command line, change your working directory to /nesi/nobackup/My_project/My_UPI /.
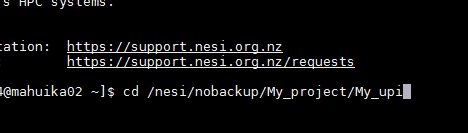
You can check your working directory by
using pwd.
3. Upload R script (and any associated files) to your working directory.
4.
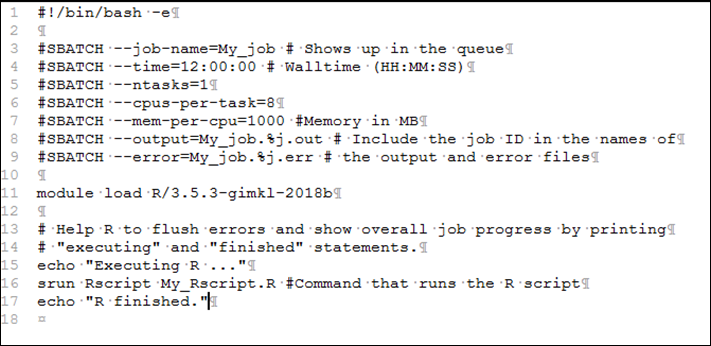
Using a text editor (e.g. Editplus), create a batch
script file and save it locally. Example files for R scripts can be found here.
Name this script with the file extension “.sl” e.g.
“My_job.sl”
 Information about ascertaining
appropriate job dimensions (e.g. memory, wall-time etc.) can be found here.
Information about ascertaining
appropriate job dimensions (e.g. memory, wall-time etc.) can be found here.
5. Upload this batch script to your working directory. You can also create your batch file directly in the Mahuika working directory using an inbuilt text editor run directly from the Mobaxterm command line. Use the command nano My_job.sl to open up the text editor. Either write or copy in the batch file text and exit the text editor with ‘ctrl + x’. This will then save your batch file My_job.sl directly in your working directory.
6. You can submit the R script job to the NeSI scheduler using sbatch My_job.sl. You should receive an output Submitted batch job 3857123. Depending on the resources required, the job might remain in the queue for a period of time. You can check the status and information about jobs you have submitted in the last day using sacct. Further information about checking your jobs can be found here.
7. When the job is complete, output and error files will be added to your working directory. You may need to refresh Mobaxterm for these files to appear.
Accessing R interactively on Mahuika
NeSI using Mobaxterm
Accessing R interactively means that you can see the R
interface and use R as you normally would on a personal computer. This can be
useful to follow the progress of a short job.
1.
Load the R module on NeSI Mahuika
using module load R/3.5.3-gimkl-2018b.
Open the R interactive session using the command R.
Commands executed will now be run in R. To exit interactive R, use the command q(). R will ask if you would like to save the workspace image. Reply y or n as required.
Instructions for running an R script interactively on NeSI Mahuika
1. Create a batch script file using a text editor (such as Editplus).
2. Copy the text in the batch script file (there is no need to upload this to your working directory, nor make the batch file in your working directory).
3.
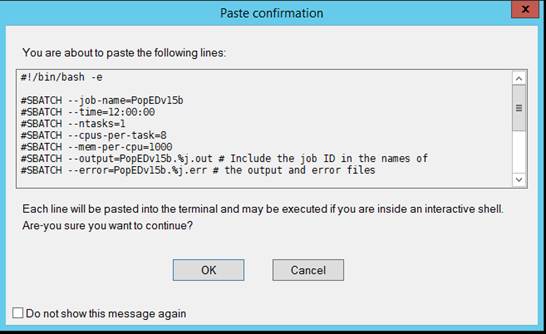
Paste the batch script text directly onto the Mobaxterm
command line. You will be presented with a popup as shown. Click OK.
Do not enter an interactive R session before pasting the batch script onto the Mobaxterm command line – this will be done automatically. The run will then start and progress will be displayed on the Mobaxterm interface.
Warning: Please note that attempting to run large R jobs interactively on Mobaxterm may result in job failure. Submit the R job as a batch file instead.